Deep learning for causal inference
This project focuses on learning deep learning models for causal inference problems, specifically estimating treatment effects on outcomes. The main objective is to explore whether a casual deep learning approach can be used for estimating drug/CNV (copy number variation) relationships in single-cell.
Notation for causal inference:
-
Observed covariates/features: $X$
-
Potential outcomes: $Y(0)$ and $Y(1)$
-
Treatment: $T$
-
Average Treatment Effect: $ATE =\mathbb{E}[Y(1)-Y(0)]$
-
Conditional Average Treatment Effect: $CATE =\mathbb{E}[Y(1)-Y(0) \mid X=x]$
The key problem in estimating treatment effects on outcome is that we only have one measurement ($Y(0)$ or $Y(1)$) for one individual. The missing outcome is counterfactual.
Here, our deep learning method focuses on learning:
- representation or latent space from the data consisting of $X$ covariates and $T$ treatment
- predict both $\hat{Y}^(0)$ and $\hat{Y}(1)$ for one individual
- use only the factual $Y(0)$ or $Y(1)$ for loss calculation using the mean squared error (MSE) and binary cross-entropy (BCE)
- calculate ATE and CATE as the following:
- $\hat{CATE}=(1-2t)(\hat(1-t)-\hat{y}(t))$
- $\hat{ATE}=\frac{1}{n}\sum_{i=1}^n\hat{CATE_i}$
Tutorial - Deep-Learning-for-Causal-Inference
The tutorial uses Tensorflow, but we use pyTorch for implementation.
The architecture of the deep learning model: Treatment Agnostic Regression Network (TARNet)
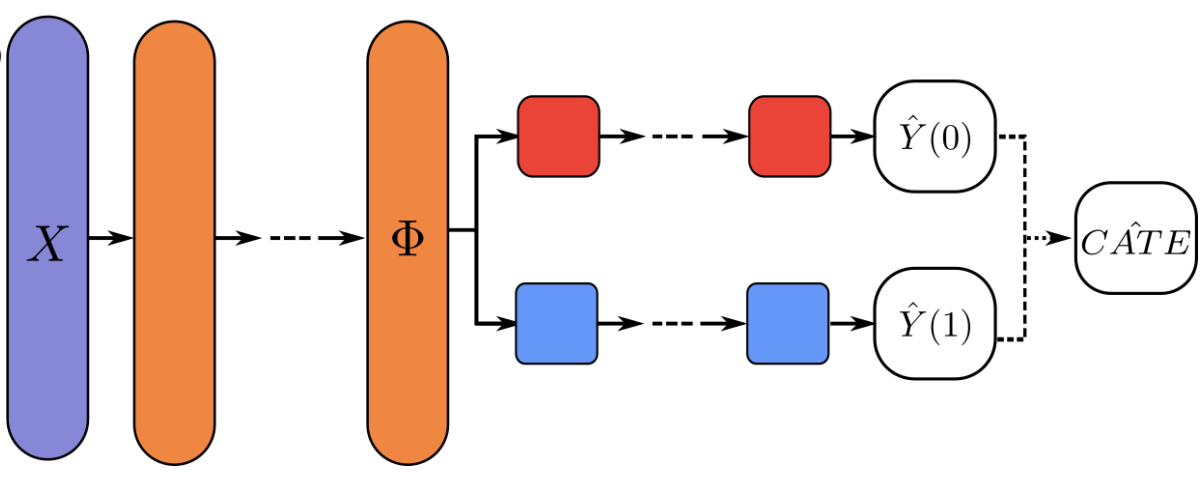
We use IHDP dataset used in the tutorial from paper Hill, 2011 to evaluate the estimation of heterogeneous treatment effects.
Download data:
wget -nc http://www.fredjo.com/files/ihdp_npci_1-100.train.npz
wget -nc http://www.fredjo.com/files/ihdp_npci_1-100.test.npz
Run Treatment Agnostic Regression Network (TARNet)
import logging
import pandas as pd
import cicnv as _cicnv
import torch
import seaborn as sns
import matplotlib.pylab as plt
from sklearn.preprocessing import StandardScaler
import numpy as np
cicnv = _cicnv.CICNV()
cicnv.data.train_data = pd.read_csv('data/ihdp_test_data.csv.gz')
cicnv.data.test_data = pd.read_csv('data/ihdp_train_data.csv.gz')
tnet = {
'out': 'output/ihdp/',
'train': {
'batch_size': 64,
'l_rate': 0.0001,
'epochs': 10000,
'enlayers': [200, 20],
'declayers': [20, 100, 1],
'device': 'cuda:1'
},
'eval': {
'batch_size': 100
},
'model_id': 'ihdp'
}
batch_size = tnet['train']['batch_size']
l_rate = tnet['train']['l_rate']
epochs = tnet['train']['epochs']
enlayers = tnet['train']['enlayers']
declayers = tnet['train']['declayers']
device = tnet['train']['device']
model_out = tnet['out']
logging.basicConfig(filename=model_out+'tnet_model.log',
format='%(asctime)s %(levelname)-8s %(message)s',
level=logging.INFO,
datefmt='%Y-%m-%d %H:%M:%S')
logging.info('tnet \n' +
'batch_size:' + str(batch_size) + '\n'
'lrate:' + str(l_rate) + '\n'
'epochs:' + str(epochs) + '\n'
'enlayers:' + str(enlayers) + '\n'
'declayers:' + str(declayers) + '\n'
'device:' + str(device) + '\n'
)
cicnv.run_tnet(batch_size,l_rate,epochs,enlayers,declayers,device)
torch.save(cicnv.tnet.model.state_dict(),model_out+'tnet.torch')
df_trainloss = pd.DataFrame(cicnv.tnet.train_loss)
df_trainloss.to_csv(model_out + 'tnet_train_loss.txt.gz')
df_testloss = pd.DataFrame(cicnv.tnet.test_loss)
df_testloss.to_csv(model_out+ 'tnet_test_loss.txt.gz')
## evaluation
cicnv.tnet.model = torch.load(model_out+'tnet.torch')
train_batch_size = cicnv.data.train_data.shape[0]
test_batch_size = cicnv.data.test_data.shape[0]
enlayers = tnet['train']['enlayers']
declayers = tnet['train']['declayers']
device = 'cpu'
y0_hat,y1_hat = cicnv.eval_tnet(cicnv.data.train_data,batch_size,enlayers, declayers,device)
cate_pred=y1_hat-y0_hat
ate_pred = torch.mean(cate_pred)
print("Estimated ATE (True is 4):", ate_pred.detach().numpy(),'\n\n')
#Estimated ATE (True is 4): 4.4657154
def load_IHDP_data(training_data,testing_data,i=7):
with open(training_data,'rb') as trf, open(testing_data,'rb') as tef:
train_data=np.load(trf); test_data=np.load(tef)
y=np.concatenate( (train_data['yf'][:,i], test_data['yf'][:,i])).astype('float32') #most GPUs only compute 32-bit floats
t=np.concatenate( (train_data['t'][:,i], test_data['t'][:,i])).astype('float32')
x=np.concatenate( (train_data['x'][:,:,i], test_data['x'][:,:,i]),axis=0).astype('float32')
mu_0=np.concatenate((train_data['mu0'][:,i], test_data['mu0'][:,i])).astype('float32')
mu_1=np.concatenate((train_data['mu1'][:,i], test_data['mu1'][:,i])).astype('float32')
data={'x':x,'t':t,'y':y,'t':t,'mu_0':mu_0,'mu_1':mu_1}
data['t']=data['t'].reshape(-1,1) #we're just padding one dimensional vectors with an additional dimension
data['y']=data['y'].reshape(-1,1)
#rescaling y between 0 and 1 often makes training of DL regressors easier
data['y_scaler'] = StandardScaler().fit(data['y'])
data['ys'] = data['y_scaler'].transform(data['y'])
return data
data=load_IHDP_data(training_data='data/ihdp_npci_1-100.train.npz',testing_data='data/ihdp_npci_1-100.test.npz')
cate_true=data['mu_1']-data['mu_0']
sns.kdeplot(data=pd.Series(cate_pred.detach().squeeze()),label='predicted')
sns.kdeplot(data=pd.Series(cate_true.squeeze()),label='true')
plt.legend()
plt.savefig(model_out+'tnet_cate_pred.png')
plt.close()
Result
Estimated ATE (True is 4): 4.4657154
And distribution plot comparing true CATE and predicted CATE:
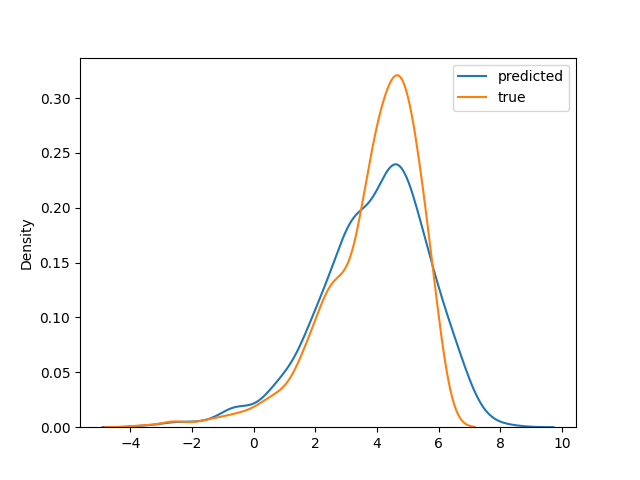
In conclusion, we tested the Treatment Agnostic Regression Network (TARNet) for estimating treatment effects using a causal inference approach. The idea of learning latent space from mixed treatment data and then predicting counterfactual treatment effects using deep neural network architecture is an interesting approach. We can expand this model for copy number variation analysis in single-cell as the following:
Estimate how drug treatment affects CNV (Copy Number Variation) using oncogene expression as input features.
- Treatment
T:-
1= Drug-treated -
0= Control (untreated)
-
- Outcome
Y:- CNV measurement (continuous or categorical)
- Covariates
X:- Gene expression levels of oncogenes (e.g., TP53, MYC, etc.)
The PyTorch implementation code used to generate the above results is available cicnv.