
Multilayer matrix factorization with neural network
This project is a proof-of-concept implementation of multilayer NMF, combining ideas from neuralNMF package and Poisson matrix factorization from ASAP package.
Key-idea Given a matrix factorization problem, i.e.
- $W \sim \theta \beta$ where,
- $W$ is
cell x genematrix - $\theta$ is
cell x factormatrix - $\beta$ is
factor x genematrix
- $W$ is
- pytorch autograd for the optimization of $\theta$ matrix
- least squares or PMF method for $\beta$ matrix optimization.
- neural network for multilayer factorization ( W -> $\theta 1$ -> $\theta 2$) where $\theta$ is new $W$ for each layer.
Data prep
We simulate a dataset consisting of 1000 cells with 2000 genes using the Poisson-Gamma model.
############### generate data
import neuralNMF
import scipy
import torch
from neuralNMF.dutil.read_write import write_h5
H,W,X = neuralNMF.generate_data(N=1000,K=10,M=2000,mode='block')
smat = scipy.sparse.csr_matrix(X)
row_names = [ str(i) for i in range(X.shape[0])]
col_names = ['c'+str(i) for i in range(X.shape[1])]
write_h5('data/sim',row_names,col_names,smat)
Run model
In this case, the matrix factorization problem is:
- $W \sim \theta \beta$ where,
- $W$ is
cell x genematrix is1000 x 2000 - $\theta$ is
cell x factormatrix- layer 1: $\theta 1$ is
1000 x 20 - layer 2: $\theta 2$ is
20 x 10
- layer 1: $\theta 1$ is
- $\beta$ is
factor x genematrix- layer 1: $\beta 1$ is
2000 x 20 - layer 2: $\beta 2$ is
2000 x 10
- layer 1: $\beta 1$ is
- $W$ is
import neuralNMF
import logging
sample = 'sim'
wdir = ''
neuralNMF.create_dataset(sample,working_dirpath=wdir)
data_mem_size = 10000
layers = [20,10]
device = 'cpu'
epochs = 300
model = neuralNMF.create_model(sample=sample,data_mem_size=data_mem_size,layers=layers,device=device,working_dirpath=wdir)
logging.info(model.net)
model.train(epochs=epochs,lr=1)
model.save()
Results
import matplotlib.pylab as plt
import seaborn as sns
import pandas as pd
import anndata as an
import neuralNMF as model
sample = 'sim'
wdir = ''
fpath = wdir+'results/'+sample
adata = an.read_h5ad(wdir+'results/'+sample+'.h5nnmf')
##plot theta
sns.clustermap(adata.uns['theta_l1'])
plt.savefig(fpath+'_theta1.png');plt.close()
sns.clustermap(adata.uns['theta_l2'])
plt.savefig(fpath+'_theta2.png');plt.close()
#plot beta
model.plot_gene_loading(adata = adata,col='beta_l1',top_n=10,max_thresh=50)
model.plot_gene_loading(adata = adata,col='beta_l2',top_n=10,max_thresh=50)
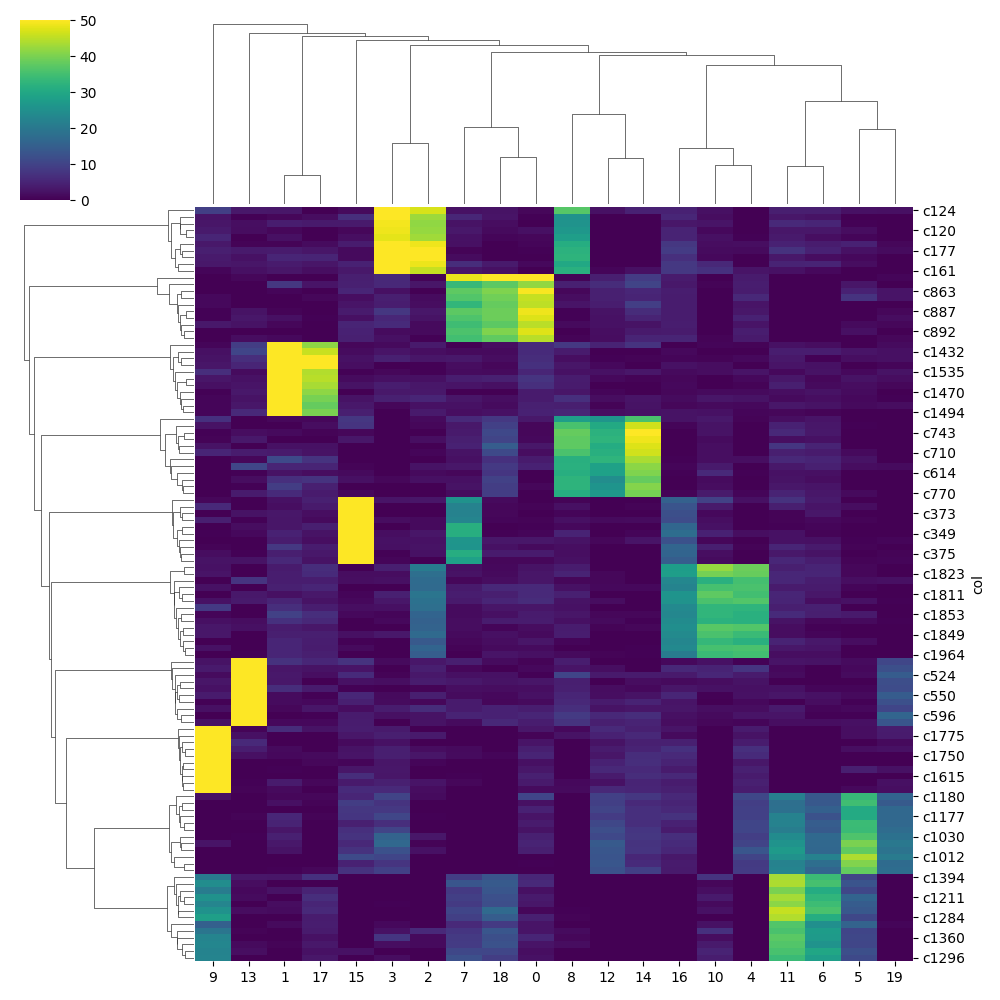
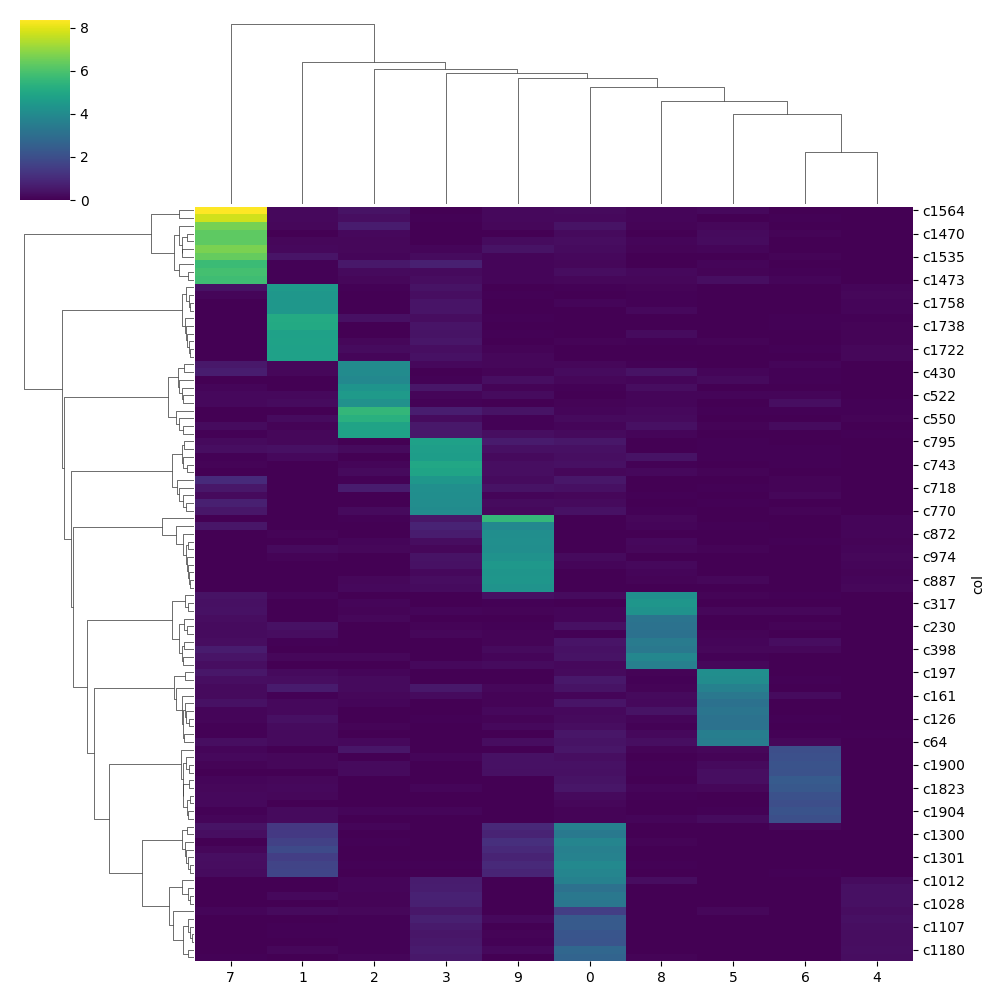
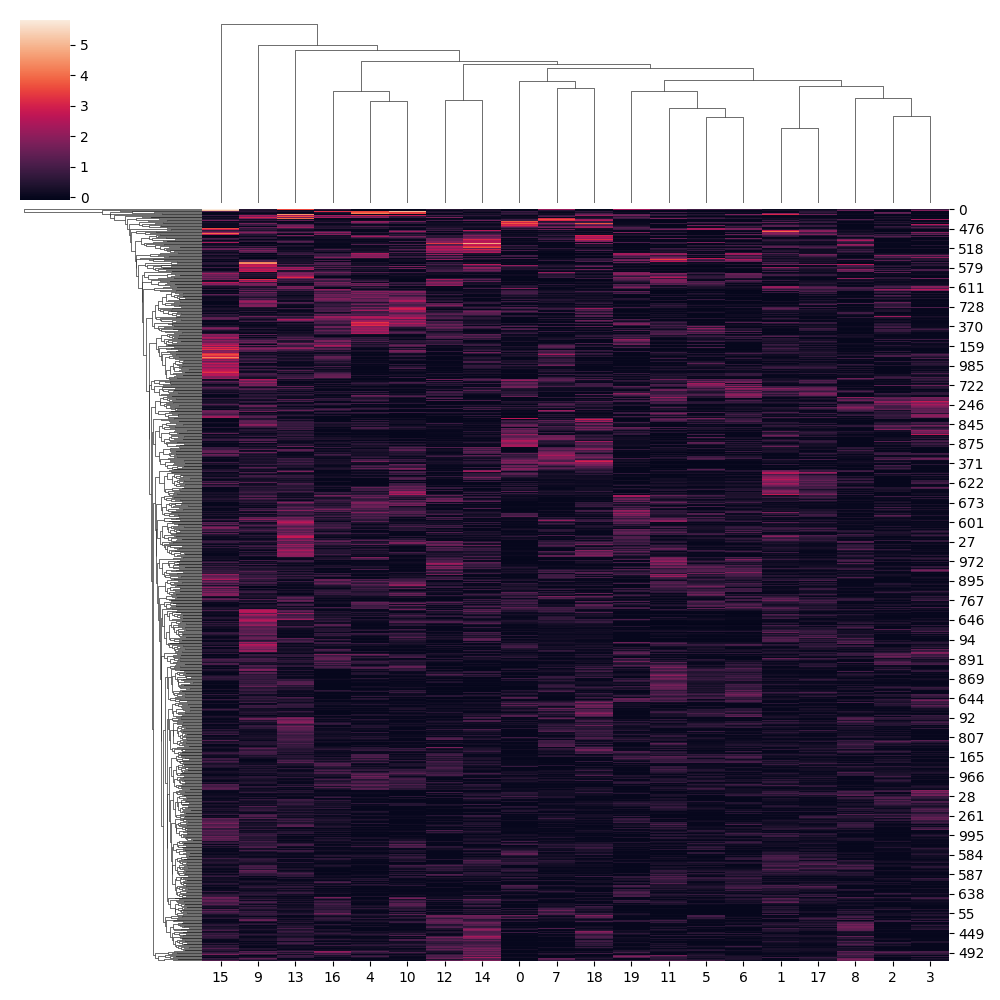
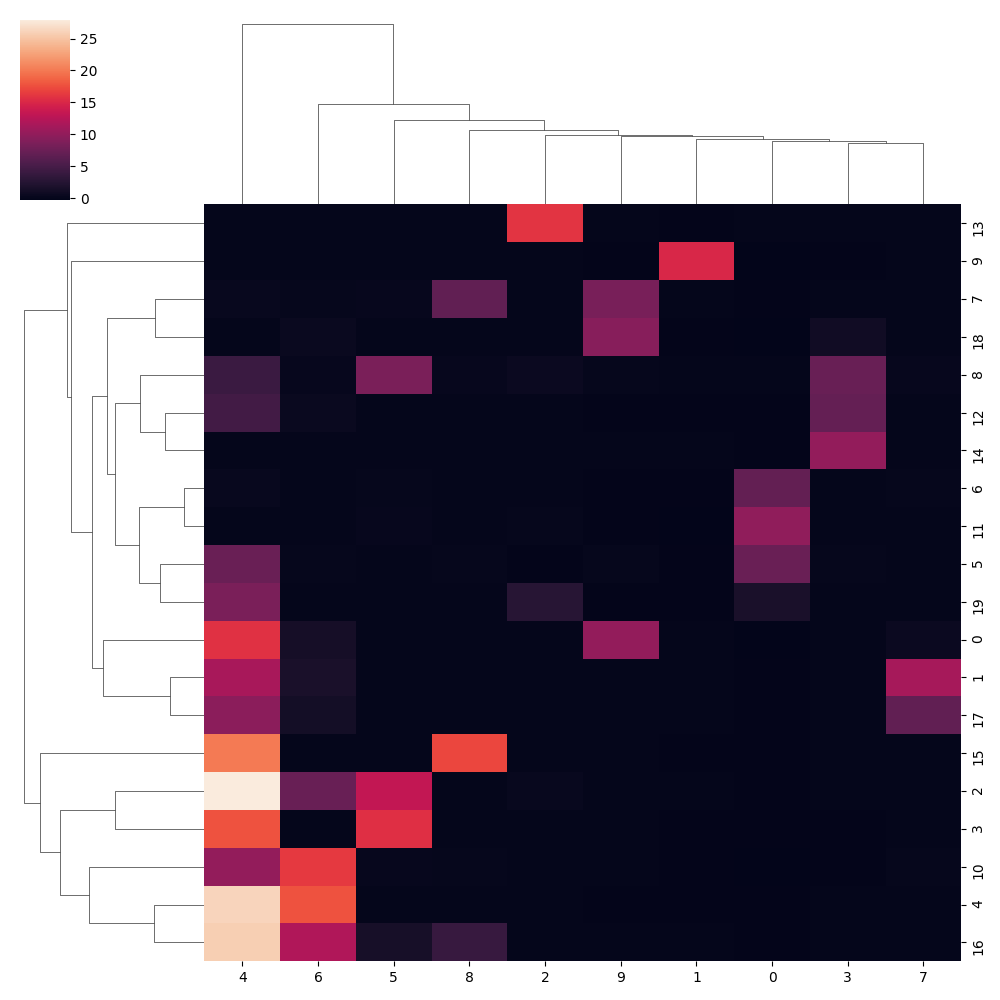
Conclusion
neuralNMF model provides a proof-of-concept modelling framework to integrate neural networks with matrix factorization techniques. To factorize $W \sim \theta \beta$, we optimized $\theta$ as neural network parameters and $\beta$ with factorization reconstruction loss. We can further refine this model to learn both $\theta$ and $\beta$ matrices as network parameters.
The project code used to generate the above results is available neuralNMF.