Attentive neural networks for biological interpretability
The biological interpretability of deep learning models has been a challenging and intriguing problem.
The primary objective of this project is to evaluate a deep learning model with robust biological interpretability.
A model, named AttnCell (where to pay “attention” in “cell” data) is focused on learning the following biological information:
- gene interaction networks
- activity of pathways in a cell
- contribution of genes in pathways
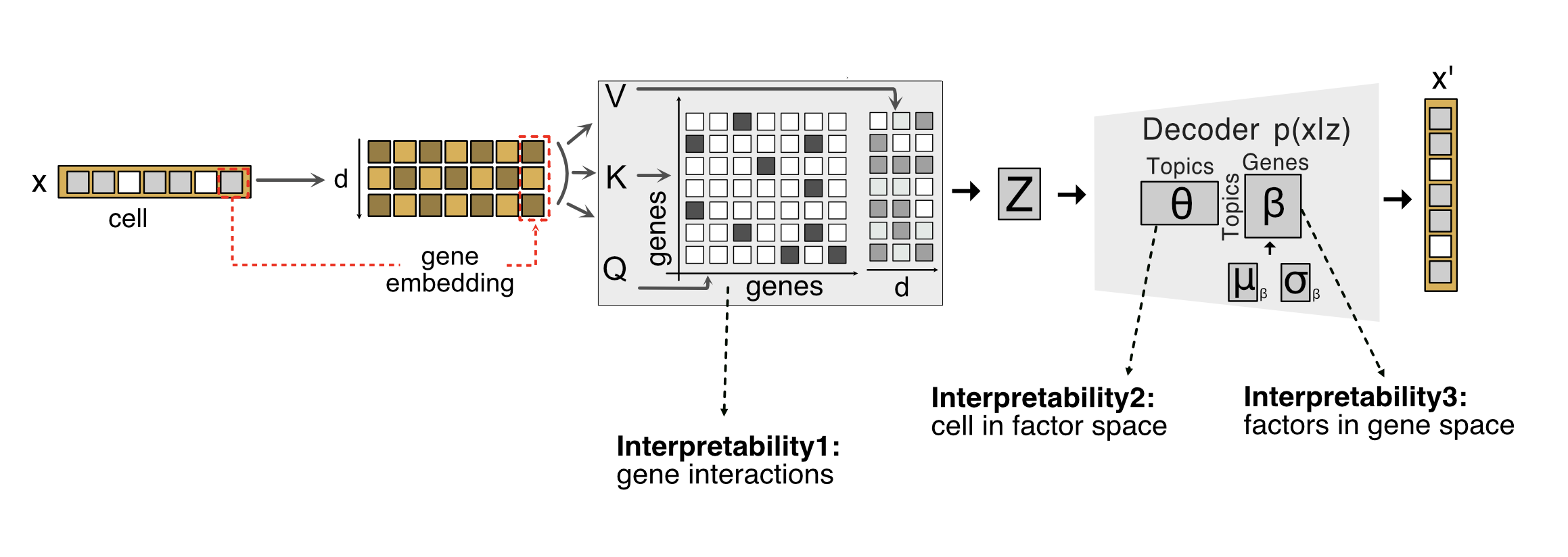
We train the model using a variational inference algorithm and maximize the evidence lower bound (ELBO). We have two losses in the model: the Kullback-Leibler (KL) loss and the Multinomial-Dirichlet likelihood loss, both of which are in the decoder network.
Preliminary results using normal pancreas data from Seurat.
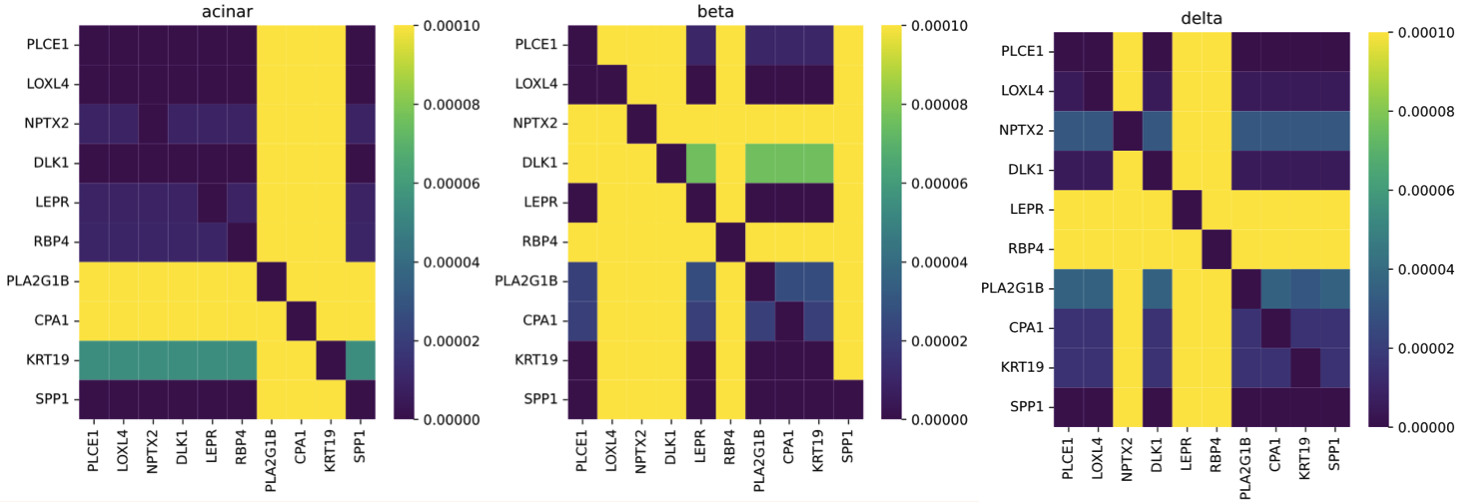
- Interpretability1: Gene interaction networks are learned by the attention module in the model. The figure illustrates examples of known marker genes, including NPTX2/DLK1 for beta cells, LEPR/RBP4 for delta cells, and PLA2G1B/CPA1 for acinar cells, in the human pancreas.
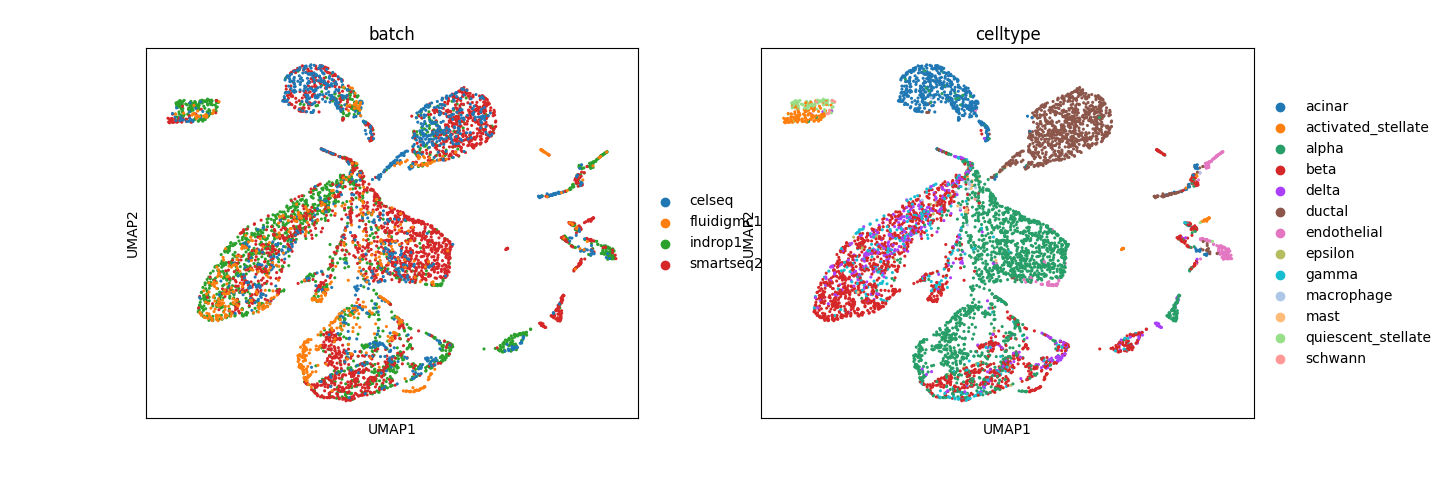
- Interpretability2: The activity of pathways in cells, which generally determines cell type identity, can be captured by analyzing the UMAP representation of cells based on the latent space learned by the model. Here, cell type-specific clusters suggest that the factors learned by the model are biologically relevant.
- Interpretability3: Contribution of genes in pathways learned as factors/topics in the model. We can further investigate different gene activity in each factor using pathway analysis.
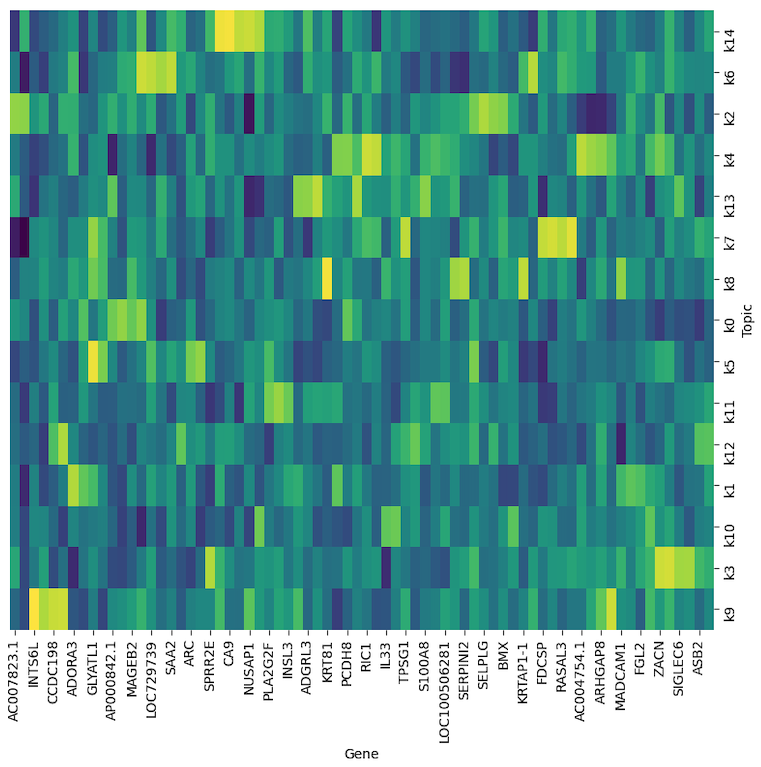
In conclusion, the results from AttnCell model are promising. We can further develop this idea to design a computational model that provides biological interpretability at multiple layers.
The code for AttnCell model is available.