Graph neural networks (GNN) for latent space decomposition
In this project, we study the Biolord paper and use a similar idea to design a new model that addresses the latent space decomposition problem.
-
Code: github repository
The authors in the paper present an interesting approach to decomposing a mixed latent space to capture label or condition-specific effects. The described deep learning model is based on a generative framework consisting of a dedicated subnetwork for each known attribute. The multiple module networks are jointly optimized.
The overview of Biolord model is:
- step 1: Generate mixed latent space
- step 2: Use label-specific subnetwork to isolate label-specific effects from mixed latent space
- step 3: Joint training with data reconstruction.
Key idea: Can we replace label-specific subnetworks with a single graph network built on a mixed space?
The updated Biolord model, named GRASP for Graph Representation Analysis for Single-cell Perturbations, consists of the following steps.
Low-dimensional space: We first aim to use any dimension reduction technique to represent high-dimensional data in a low-dimensional space. This space is used to generate an attribute-specific graph.
Attribute-specific graphs: Next, in a low-dimensional space, we identify similar cells from different groups. This approach will construct a cell-cell similarity graph in adjacent matrix format such that each cell has an edge with similar cells that belong to different labels. For example, if we have batch and cell-type labels, we will generate batch and cell-type-based graphs. In a batch-based graph, edges are constructed between similar cells from different batches (most likely from the same cell type). Similarly, in a cell type-based graph, edges are constructed between similar cells from different cell types (most likely from the same batch).
Why graphs? First, a single graph in mixed space replaces attribute-specific multiple modules presented in Biolord. We can use the graph repeatedly to construct attribute-specific representations. This will provide scalability to the model. Second, when we use attribute-specific graphs, we can learn shared effects specific to the attribute. We can guide the shared effect using GNN to generate attribute-specific latent space.
Simplified training: In Biolord, we have multiple attribute-specific losses, but in GRASP, we have only two losses-
- reconstruction loss and
- alignment loss to encourage independence among attribute-specific factors.
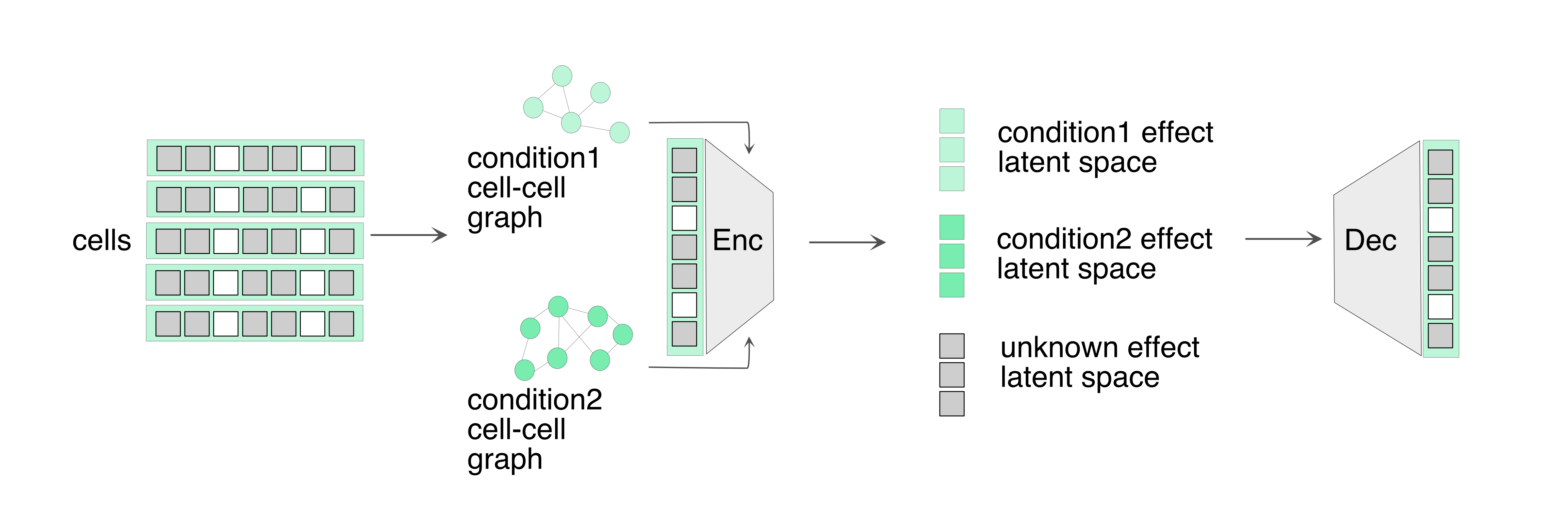
GRASP model:
- Here, we use batch and cell type labels as two attribute labels.
- Pre-train steps:
- Use any latent space representation model (such as PCA) to obtain z_pca
- Generate a graph based on z_pca space and project it to batch space and group space
- GRASP training :
- input: raw data and two graphs in batch space and group space
- model :
- Encode raw data to z_mix
- Capture batch effect z_batch using GNN(z_mix,batch space graph)
- Capture group effect z_group using GNN(z_mix,group space graph)
- Isolate z_unknown from FCN (z_mix, [z_batch + z_group])
- Reconstruct data using z_batch + z_group + z_unknown
- Discriminator learning for batch and group effect
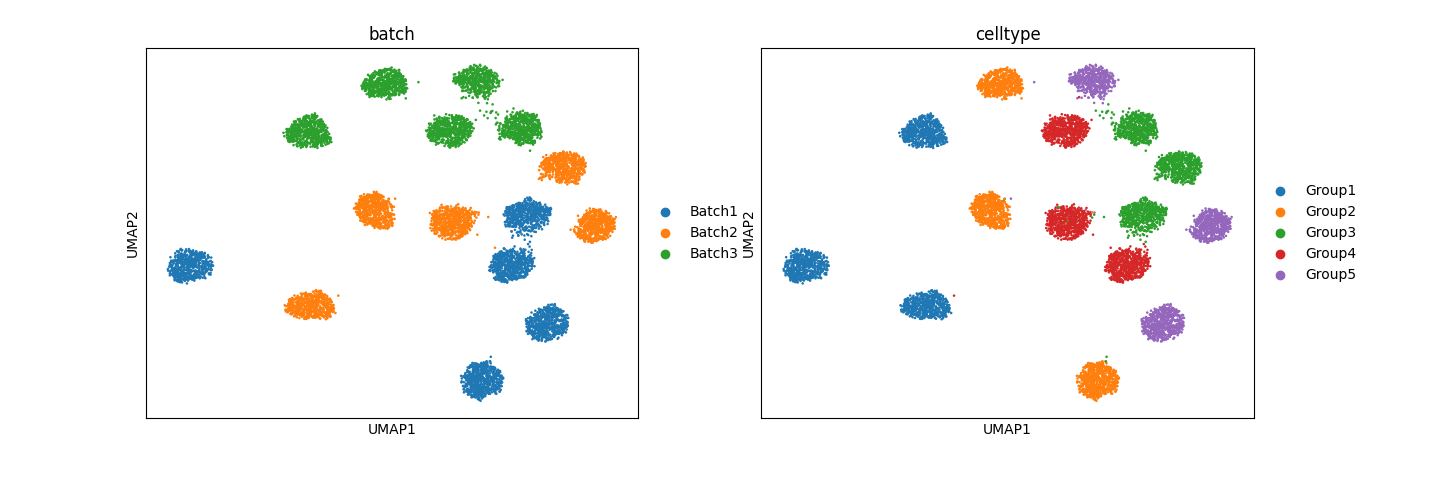
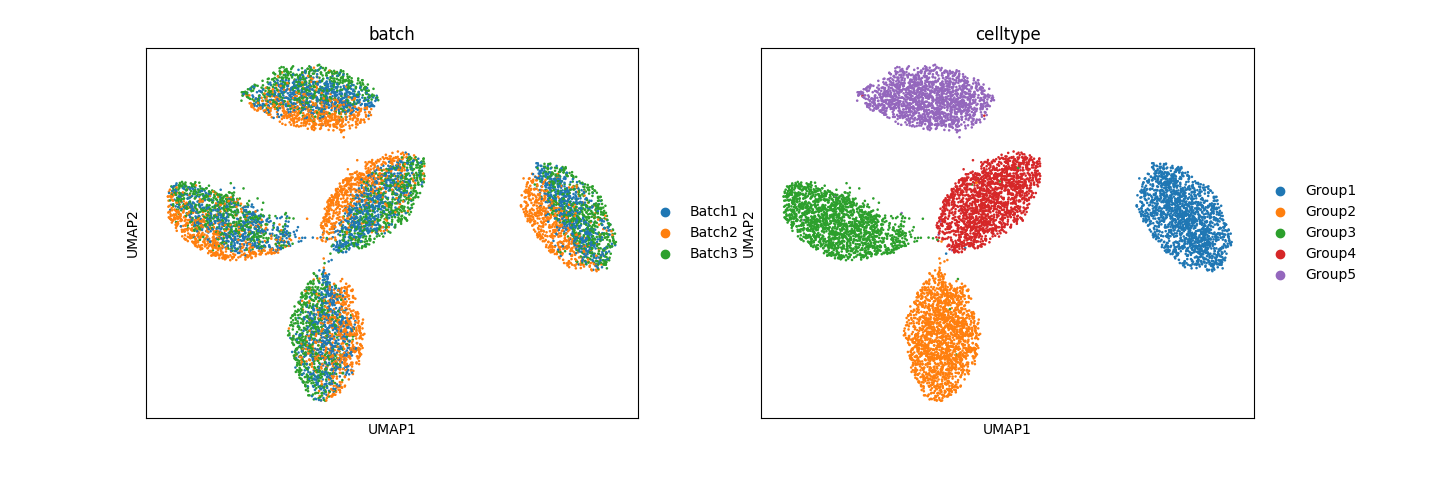
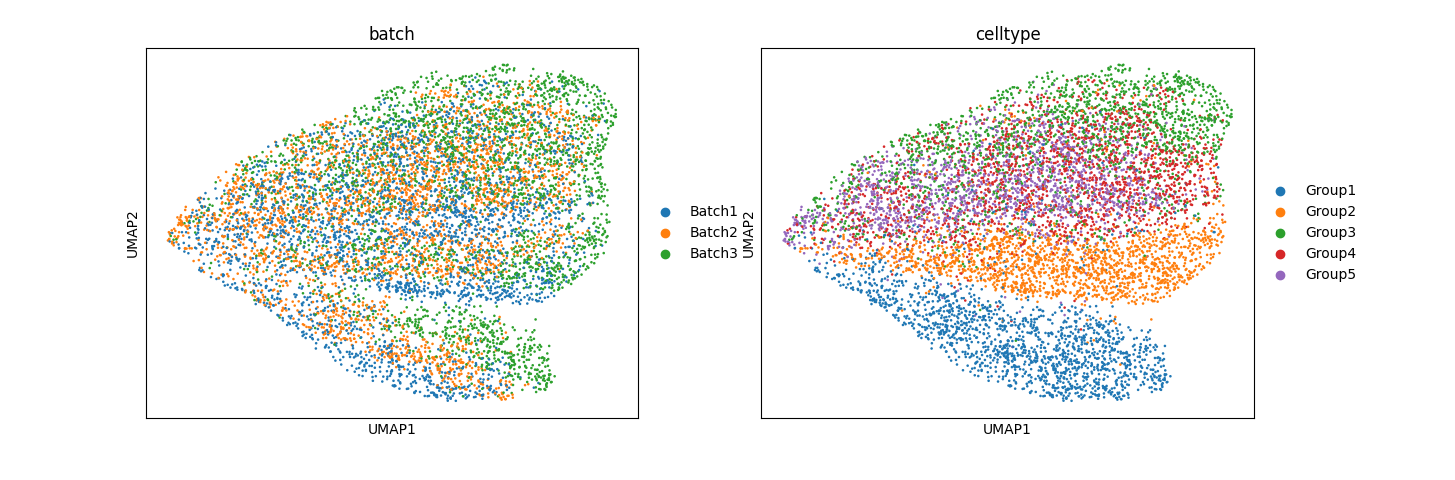
Preliminary results for simulation data:
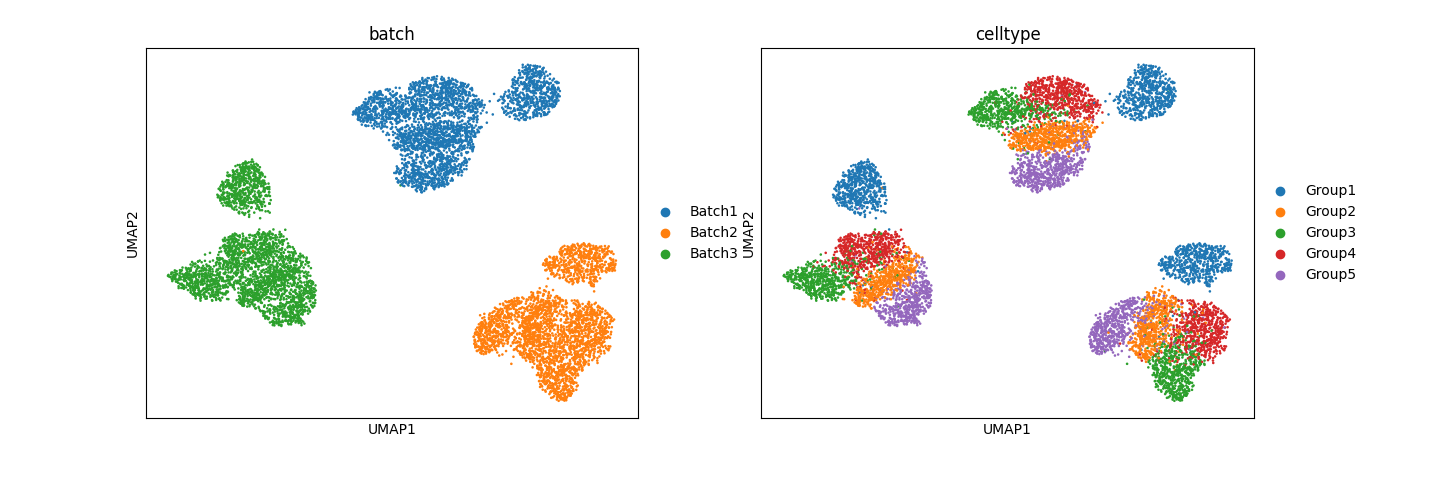
Preliminary results for normal pancreas data from Seurat:
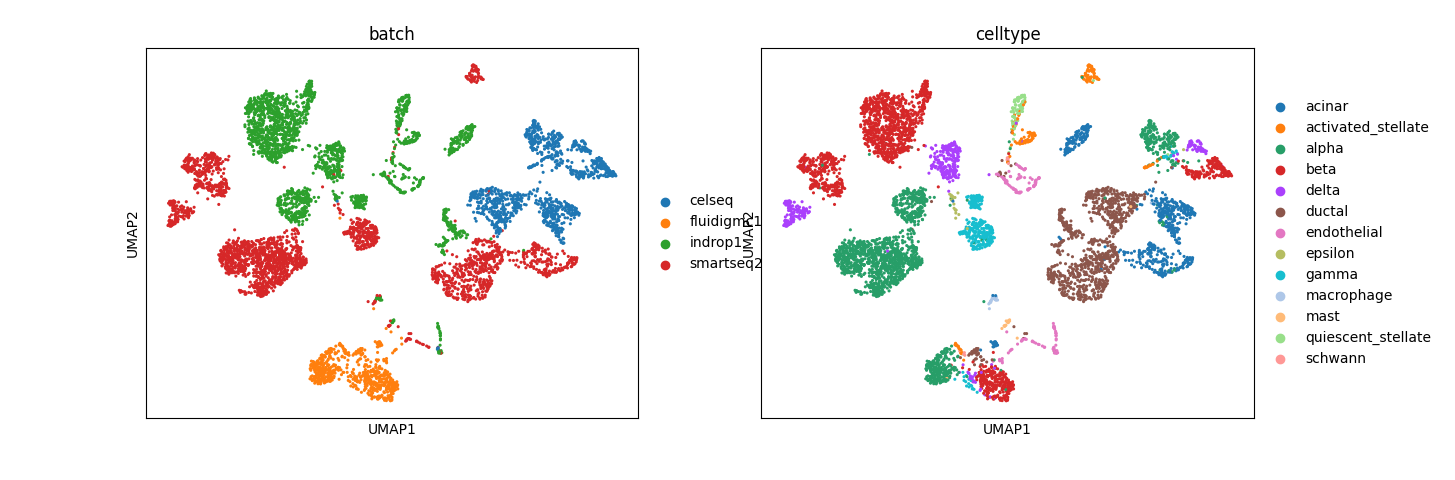

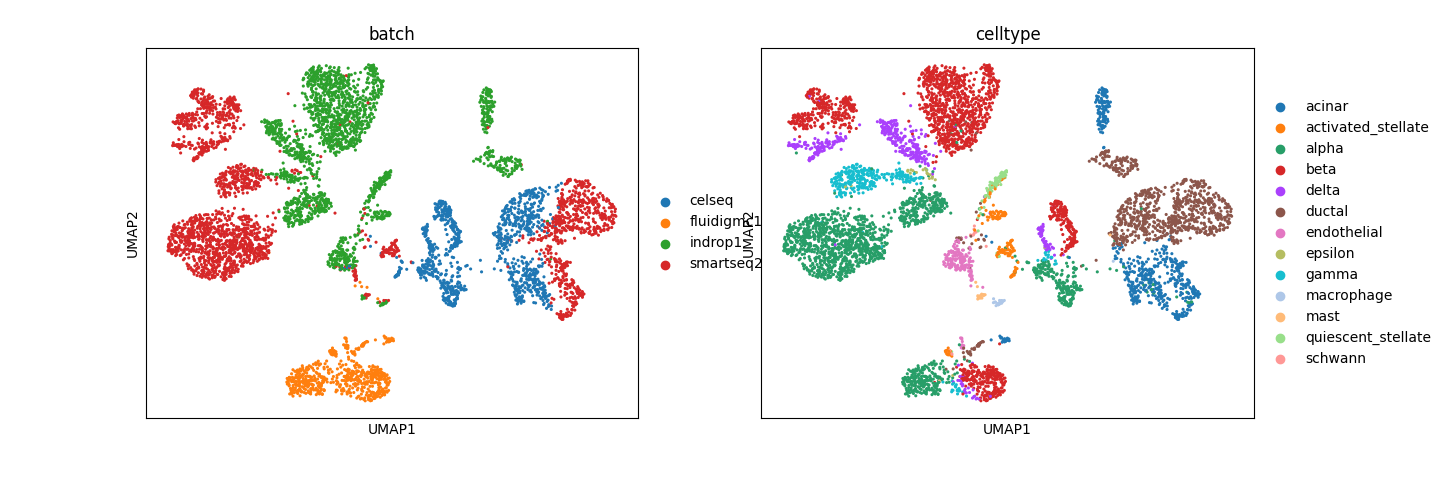
The results, especially from simulation data, are promising. In mixed space, we observe that the batch effect is dominant, followed by the cell type effect, which generates unique clusters. Attribute-specific representations exhibit distinct clusters that capture the effects of each attribute. However, the results from the normal pancreas data are not convincing and suggest that more work is needed to refine the model.
The project code used to generate the above results is available GRASP.